2014
How duplicated transcription regulators can diversiFy to govern the expression of nonoverlapping sets of genes.
Perez, J.C., Fordyce, P.M., Lohse, M.B., Hanson-Smith, V., DeRisi, J.L., & Johnson, A.D. Genes & Development (2014).
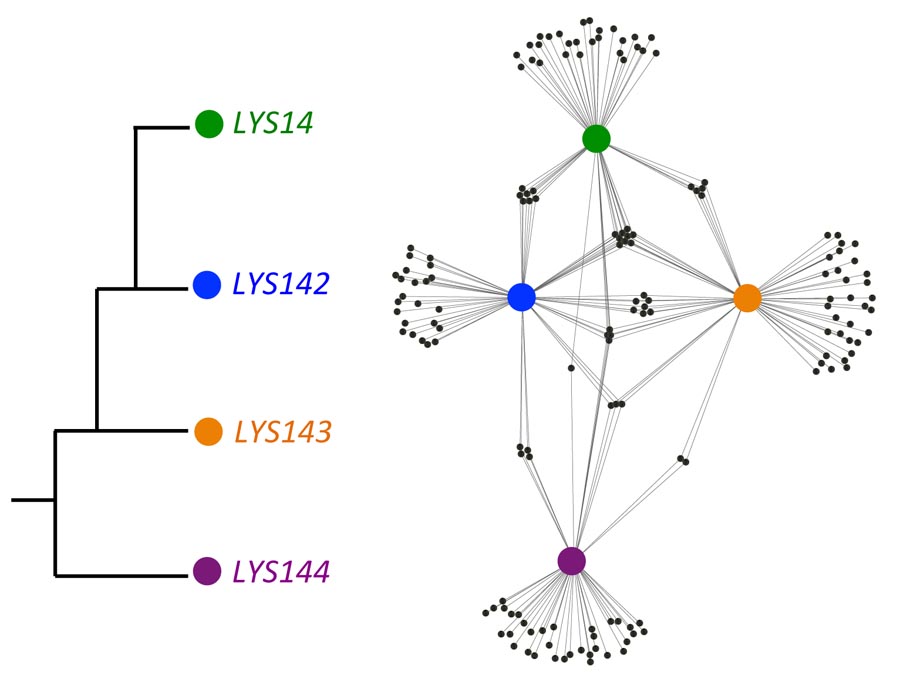
Gene expression is regulated primarily by transcription factor proteins that bind to DNA regulatory sequences to activate or repress transcription. Evolutionary changes in these binding interactions are thought to be driven primarily by changes in DNA regulatory sequence. Here, we show that small changes in transcription factor specificity can also rewire networks by investigating how 4 related transcription factors in Candida albicans have evolved to recognize largely distinct targets.
Hydraulic expulsion of Tumbu fly larvae
Dybbro, E., Fordyce, P., Ponte, M., & Arron, S.T. JAMA Dermatol. (2014).
Furuncular myiasis is rarely observed outside of endemic areas, which may hamper accurate diagnosis. This paper reports two cases of Cordylobia anthropophaga larval removal from two patients recently returned from travel in southwestern Uganda using a larval expulsion technique.
2013
Structure of the transcriptional network controlling white-opaque switching in Candida albicans.
Hernday, A.D., Lohse, M.B.*, Fordyce, P.M.*, Nobile, C.J., DeRisi, J.L., & Johnson, A.D. Molecular Microbiology (2013) (* denotes equal authorship).
Candida albicans is the most common human fungal pathogen. It exists in one of two genetically identical but phenotypically distinct states (white and opaque) that are stably inherited over many generations. Earlier work had identified key transcriptional regulators involved in this process; however, the network remained incomplete. We identify several additional transcription factors involved in white-opaque switching and use detailed studies of their in vitro binding specificities to reveal the full network structure.
Microfluidic affinity and ChIP-seq analyses converge on a conserved FOXP2-binding motif in chimp and human, which enables the detection of evolutionarily novel targets.
Nelson, C.S., Fuller, C.K., Fordyce, P.M., Greninger, A.L., Li, H., & DeRisi, J.L. Nucleic Acids Research (2013).
FOXP2 (often nicknamed the "human speech gene") is a forkhead transcription factor required for human speech. A single amino acid difference between the human and chimp versions of the protein has been shown to be crucial for function, but whether this difference affects transcription factor binding specificity or downstream activation remained unknown. Using microfluidic affinity assays, we show that the binding specificity for the human and chimp FOXP2 proteins is identical, and use this newfound information to predict novel targets in humans.
Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains.
Lohse, M.B., Hernday, A.D., Fordyce, P.M., Noiman, L., Sorrells, T.R., Hanson-Smith, V., Nobile, C.J., DeRisi, J.L., & Johnson, A.D. PNAS (2013).
Most efforts to identify transcription factors in newly sequenced genomes rely on searches for proteins similar to previously characterized transcription factors. Here, we identify a new sequence-specific DNA-binding transcription factor (Wor3) in Candida albicans with no homology to previously identified nucleic acid binding proteins. Using a combination of microfluidic affinity assays and chromatin immunoprecipitation assays, we derive a binding specificity for this new protein family and predict direct in vivo targets. These results suggests that many more divergent transcription factors may exist that previously thought.
2012
Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses.
Fordyce, P.M., Pincus, D., Kimmig, P., Nelson, C.S., El-Samad, H., Walter, P., & DeRisi, J.L. PNAS (2012).
Basic leucine zipper (bZIP) transcription factors are thought to be among the most structurally simple transcription factors. Hac1, a bZIP transcription factor for budding yeast, plays a central role in allowing cells to sense and respond to the accumulation of unfolded proteins during stress. Despite this important role, however, there were conflicting reports about the identity of the DNA sequences bound by Hac1. Using a combination of microfluidic affinity analyses and a novel genetic screen, we show that Hac1 actually recognizes multiple sequences using a part of the protein outside of the canonical DNA binding domain. These results suggest that even structurally simple transcription factors may have more complex behaviors than previously thought, with implications for increasing regulatory flexibility in vivo.
Programmable microfluidic synthesis of spectrally encoded microspheres.
Gerver, R.E.*, Gomez-Sjobert, R.*, Baxter, B.C.*, Thorn, K.S.*, Fordyce, P.M.*, Diaz-Botia, C.A., Helms, B.A., & DeRisi, J.L. Lab on a Chip (2012) (* denotes equal authorship; author order was determined by random draw).
The ability to perform multiple biological assays in parallel using very small amounts of sample is key to improving disease diagnostics. Key to this multiplexing is the ability to mix multiple analytes together and still be able to distinguish them from one another. Here, we present a novel microfluidic system for creating spectrally encoded beads based on the ratiometric incorporation of different lanthanide nanocrystals. Using 3 different lanthanide ions (Europium, Samarium, and Dysprosium), we demonstrate automated production of 24 different codes, with the potential to make much larger code sets.
Systematic characterization of feature dimensions and closing pressures for microfluidic valves produced via photoresist reflow.
Fordyce, P.M., Diaz-Botia, C.A., DeRisi, J.L., & Gomez-Sjoberg, R. Lab on a Chip (2012).
Producing fully closing valves on two-layer microfluidic devices with tall channels is difficult because the final height at the end of the fabrication process depends on the dimensions of the valve itself. In addition, it remains difficult to predict valve closing pressures. This paper streamlines device design by determining a functional relationship between final valve height and valve geometries, and provides a model describing how closing pressure depends on valve dimensions and channel height. An associated software tool to aid in device design can be found here.
2011
Integrating systems biology data to yield functional genomics insights.
Fordyce, P., & Ingolia, N. Genome Biology (2011).
A report on the EMBO Conference "From Functional Genomics to Systems Biology" held at the EMBL Advanced Training Centre, Heidelberg, Germany, 13-16 November 2010.
2010
De novo identification and biophysical characterization of transcription factor binding with microfluidic affinity analysis.
Fordyce, P.M.*, Gerber, D.*, Tran, D., Zheng, J., Li, H., DeRisi, J.L., and Quake, S.R. Nature Biotechnology (2010) (* denotes equal authorship).
Predicting the DNA sequences bound by individual transcription factors from sequence or structure is difficult. Experimental data are required to link individual transcription factors with their cognate regulatory sequences; however, most current techniques provide only qualitative information about specificity and require large amounts of sample. Here, we present a microfluidic technique for making quantitative measurements of binding affinities between a single transcription factor protein and up to 4000 distinct DNA sequences. Using these data, we generate simple models of transcription factor specificity that can be used to predict in vivo regulatory targets. To validate this new technique, we measure 28 different transcription factor proteins from budding yeast.
2008
Advances in surface-based assays for single molecules.
Fordyce, P.M.*, Valentine, M.T.*, & Block, S.M. in "Single-Molecule Techniques: A Laboratory Manual" (Cold Spring Harbor Monograph Series) (2008) (* denotes equal authorship).
Single-molecule assays depend critically on surface treatments that ensure both robust and specific attachment of molecules of interest and successful repulsion of molecules that can interfere with measurements. This chapter provides a review of surface treatments useful for single-molecule assays, with a focus on novel polymers that can be used to tune surface charges or form dense repulsive coatings. In addition, the end of the chapter provides detailed protocols for creating each surface.
2006
Eg5 steps it up!
Valentine, M.T., Fordyce, P.M., & Block, S.M. Cell Division (2006).
Recent work has demonstrated that single human mitotic kinesin Eg5 dimers can move processively and support load in vitro. This review explores recent nanomechanical evidence surrounding Eg5 motion and mechanochemistry.
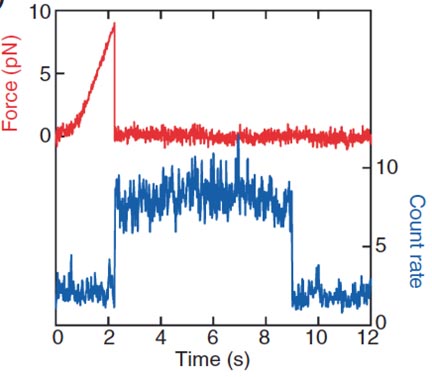
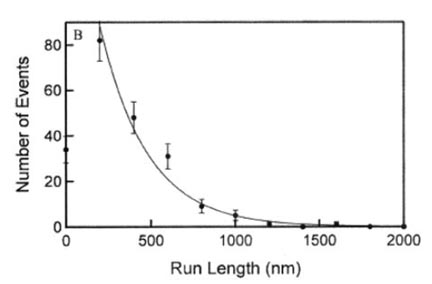
Individual dimers of the mitotic kinesin Eg5 step processively and support substantial loads in vitro.
Valentine, M.T.*, Fordyce, P.M.*, Krzysiak, T.C., Gilbert, S.P., & Block, S.M. Nature Cell Biology (2006).
Human Eg5 is a kinesin family motor protein required for spindle assembly and maintenance during mitosis. Despite its role in such a critical task, little is known about how it accomplishes this function. Here, we employ a high-precision optical trapping assay in combination with novel polymer-coated surfaces to directly probe the mechanochemistry of individual Eg5 dimers. We find that individual Eg5 motors are processive, with short run lengths of about 8 8-nm steps, on average. In comparison with conventional kinesin motors, Eg5 is slower and less sensitive to force, possibly reflecting the distinct demands of mitotic spindle assembly.
2004
Simultaneous, coincident optical trapping and single-molecule fluorescence.
Lang, M.J.*, Fordyce, P.M.*, Engh, A.M., Neuman, K.C., & Block, S.M. Nature Methods (2004).
This paper describes the construction of the first microscope capable of simultaneous, spatially coincident optical trapping and single-molecule fluorescence. To demonstrate the capabilities of the instrument, we unzip and shear apart dye-labeled DNA duplexes and establish that changes in fluorescence occur simultaneously with mechanical transitions. These measurements establish that rupture forces depend critically on the direction of loading.
2003
Combined optical trapping and single-molecule fluorescence.
Lang. M.J., Fordyce, P.M., & Block, S.M. Journal of Biology (2003).
This paper describes the initial construction of a microscope capable of simultaneous optical trapping and single-molecule fluorescence, and presents preliminary data showing mechanical rupture and loss of fluorescence from individual fluorescently labeled DNA duplexes.
Stepping and stretching: how kinesin uses external strain to step processively.
Rosenfeld, S.S., Fordyce, P.M., Jefferson, G.M., King, P.H., & Block, S.M. Journal of Biological Chemistry (2003).
Individual kinesin motors can travel long distances without detaching from microtubules, posing a puzzle: how do the two kinesin heads coordinate their activities to prevent simultaneous detachment? Using a combination of single-molecule assays and stopped-flow kinetics, this paper presents evidence that this coordination results from rearward strain on the forward head when both heads are bound.
2001
Search for the decay KL -> pi0e+e-.
Alavi-Harati A., et al. Physical Review Letters (2001).
Measurement of the branching ratio of the decay KL -> e+e-gammagamma.
Alavi-Harati A., et al. Physical Review D (2001).