High-throughput and quantitative biochemistry, biophysics, and single-cell assays
Cellular function and organismal homeostasis are governed by molecular interactions. Protein-DNA binding interactions are essential for regulating gene transcription and translation, dense networks of protein-protein and protein-peptide interactions further regulate cellular function, and enzymes make possible all of the chemical transformations essential to metabolism and signaling. Our goal is to understand, and eventually engineer, these complex processes by building and testing biophysical models of how the molecules that drive these processes work. To do so, an essential first step is to obtain the necessary quantitative measurements of the fundamental kinetic and thermodynamic constants of these molecular interactions and catalytic processes—the “universal language” needed to describe and ultimately predict function. In our lab, we use microfluidics and extensive hardware automation to perform these quantitative measurements at an unprecedented scale.
High-throughput, quantitative biochemistry & biophysics using valved Devices
Michael Hayes (Graduate Student, Genetics): Investigating how intrinsically disordered regions of transcription factors affect DNA binding
Peter Suzuki (Graduate Student, Bioengineering): Measuring weak protein-protein interactions with STAMMPPING.
Micah Olivas (Graduate Student, Genetics): Using HT-MEK to map properties of enzymatic catalysis to protein sequence landscapes
Matt DeJong (Graduate Student, Chemical Engineering): Single-molecule mechanobiology
Yujia Bian (Graduate Student, Chemical Engineering): Single-molecule identification of protein catch bonds
Maya Sheth (Graduate Student, Bioengineering): Understanding transcription factor/co-activator interactions
Jessica Karaguesian (Graduate Student, Bioengineering): Quantifying protein-protein interactions to train predictive ML models
Fanny Sunden (Research Associate, Biochemistry): Developing platforms for protein stability measurements
Patrick Almhjell (Postdoctoral Fellow): Using genetic code expansion to dissect fundamental properties of catalysis
Shawn Costello (Postdoctoral Fellow): Local and global protein stability measurements
Albert Lee (Postdoctoral Fellow): Characterization of SHP2 allelic variants for precision medicine via HT-MEK
Renee Hastings (Graduate Student, Biophysics): Investigating protein sequence determinants of nucleic acid binding specificity
Eliel Akinbami: (Graduate Student, Bioengineering): Exploring sequence-function space for PafA orthologs
Karl Krauth (Postdoctoral Fellow): Quantifying protein-protein interactions to train predictive ML models
Alan Su (Graduate Student, Chemical & Systems Biology): Probing the biophysical determinants of natural selection in Drosophila proteins
MRBLEs: Microspheres with Ratiometric Barcode Lanthanide Encoding
Caroline Horn (Lab Manager): Rapid detection of pathogens
Minsung Cho (Graduate Student, Biophysics): Multiplexed binding assays
Gabriela Lomeli (Postdoctoral Fellow): Quantifying protease specificity and cleavage rates
Micah Lawrence (Graduate Student, Bioengineering): Automation of MRBLEs generation
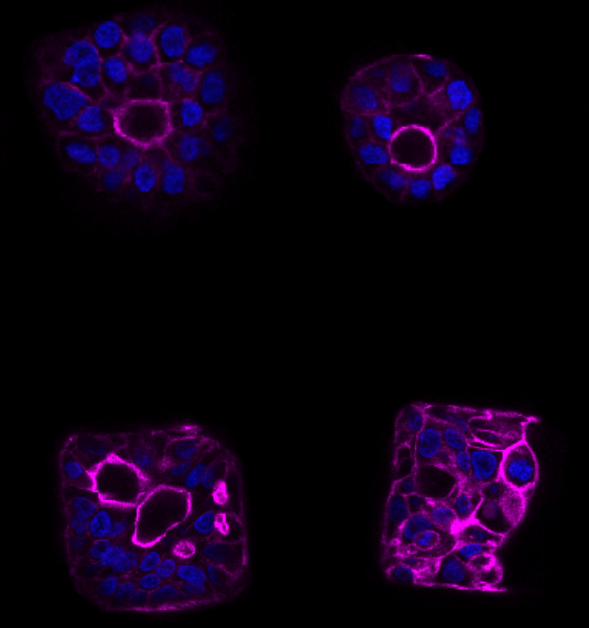
DROPCEPTION: single-cell biology using sortable double emulsion droplets
Conor McClune (Postdoctoral Fellow): Single cell technologies to dissect plant biosynthetic pathways
Samuel Thompson (Postdoctoral Fellow): Engineering proteins for non-aqueous environments
Daria Passow (Graduate Student, Biophysics): Bead and droplet assays for ultra-high-throughput protein characterization
MICROWELLS FOR HIGH-THROUGHPUT SINGLE-CELL PHENOTYPING
Collaborators: Kasper Karlsson (postdoctoral fellow, Curtis lab, Genetics).
If you’d like to join us for Group Meeting we’d love to have you!
Here’s our Schedule Document!


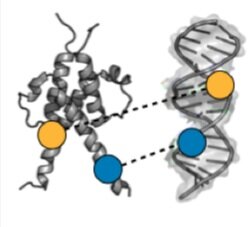
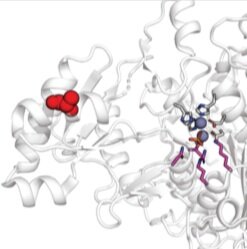
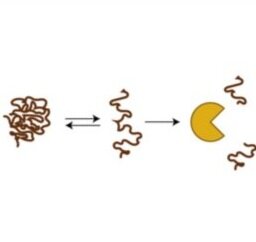
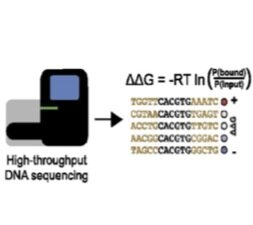
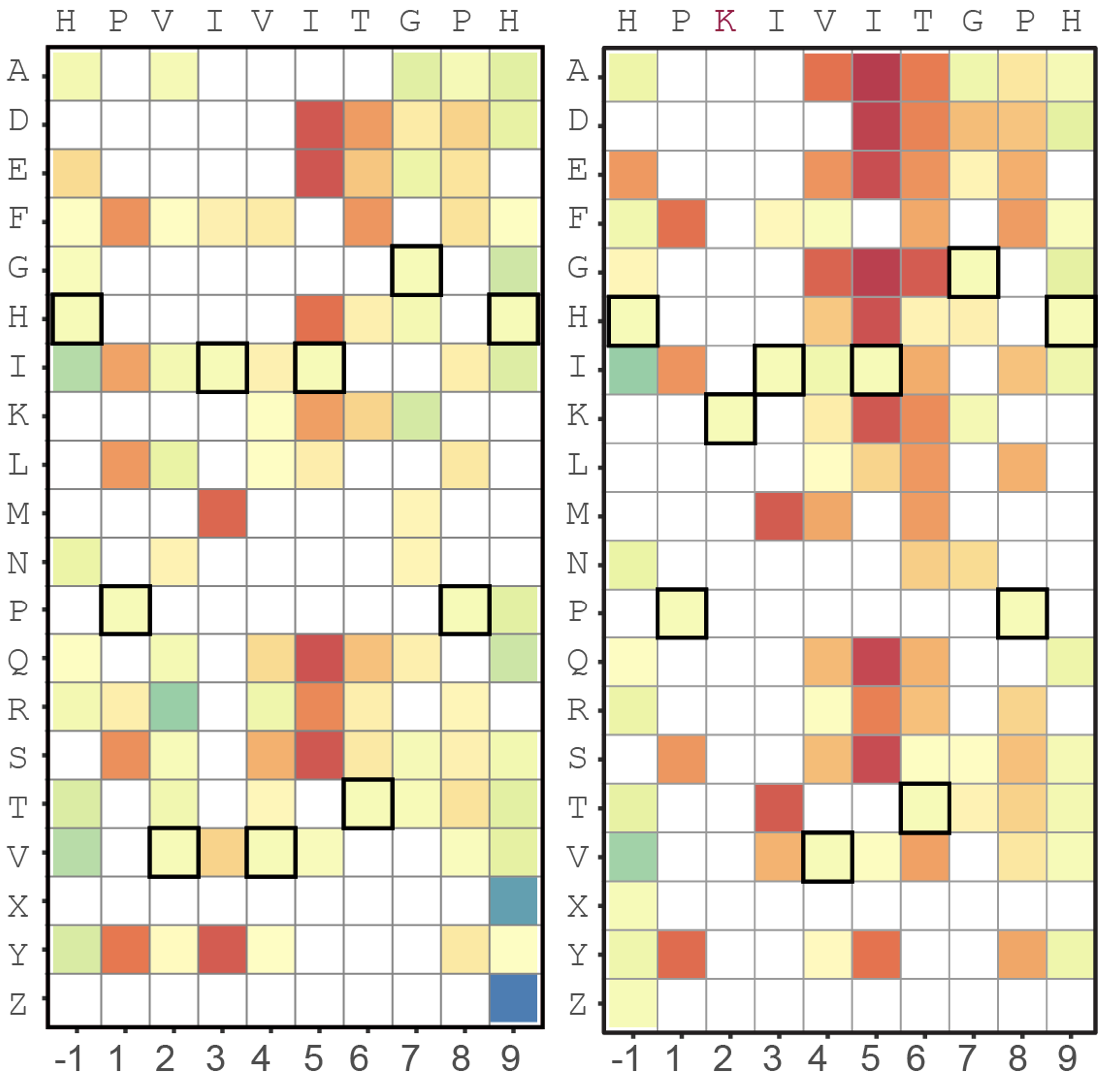
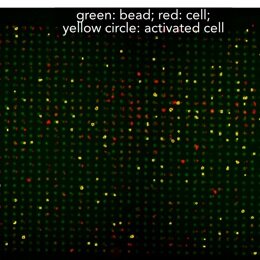

Array-based multiplexing experiments (MITOMI and HT-MEK) employ microfluidic devices containing 1,568 valved reaction chambers aligned to printed DNA arrays. We are currently using these devices to better understand how transcription factors find and bind their genomic targets to regulate gene expression, as well as to understand how enzymes achieve their extraordinary catalytic efficiency and substrate specificity.