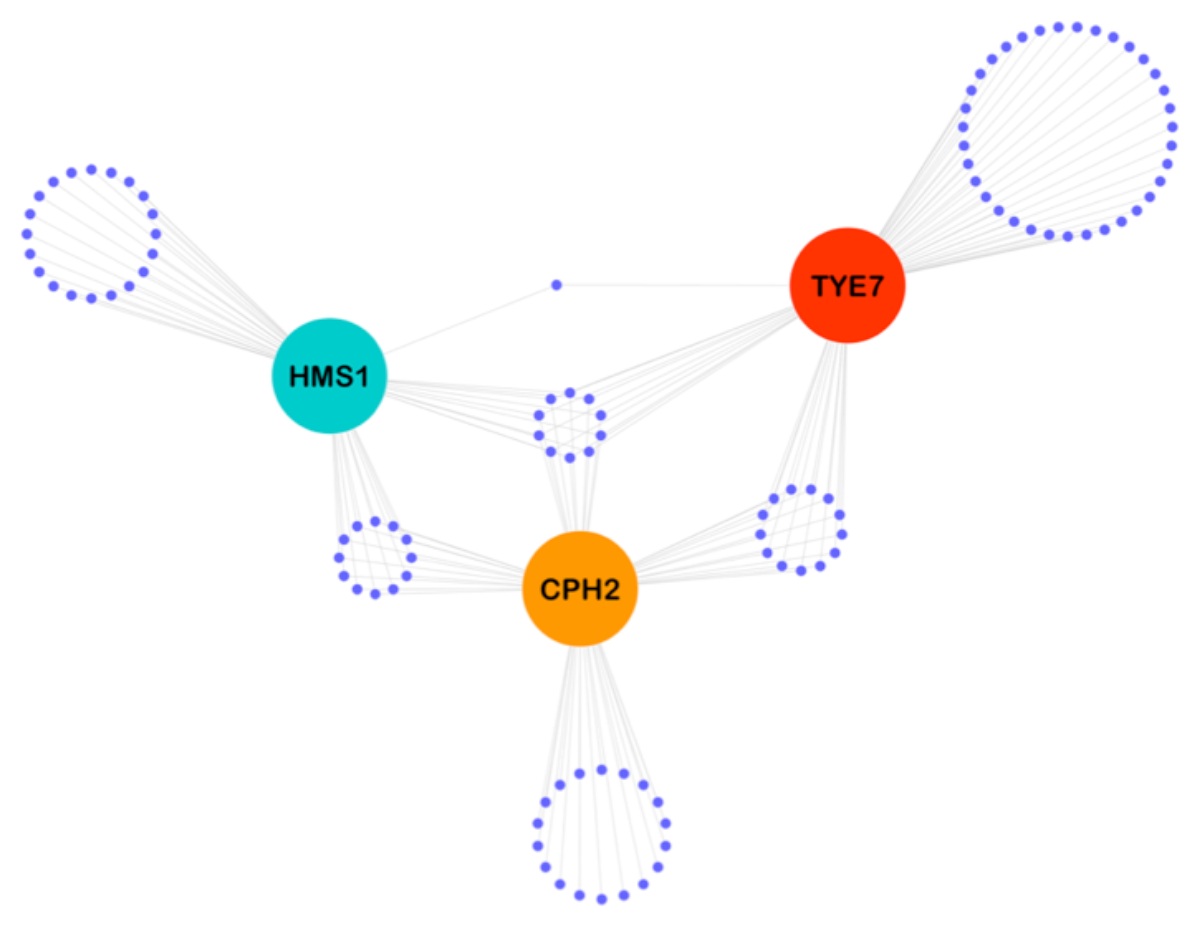
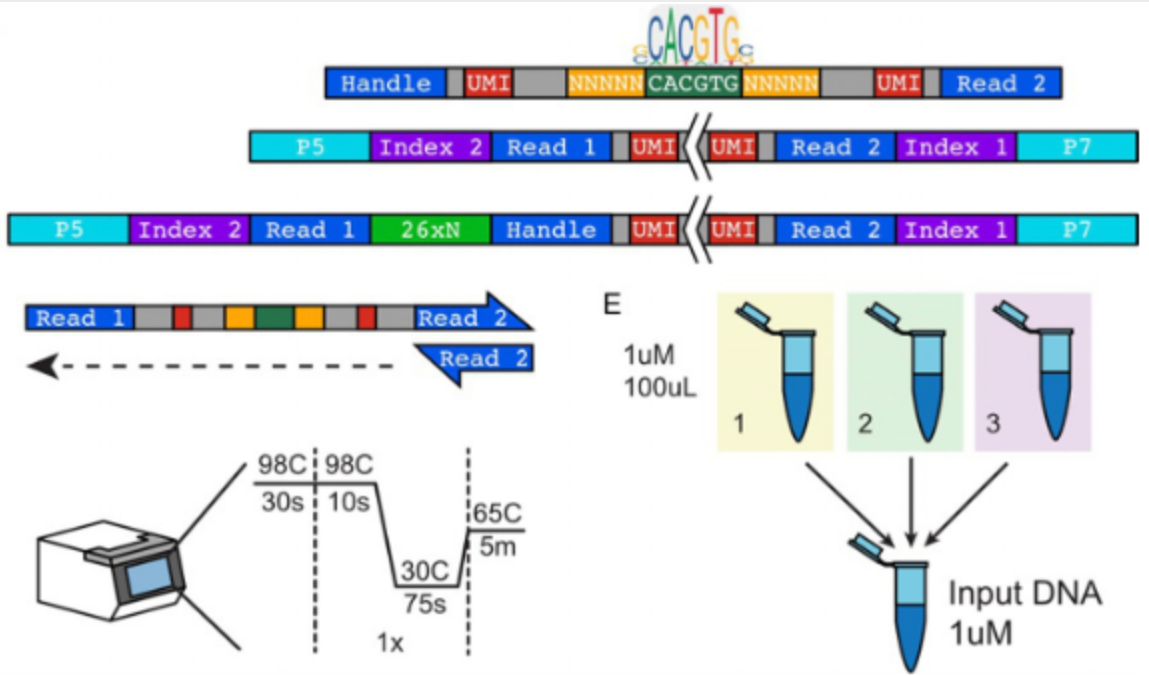
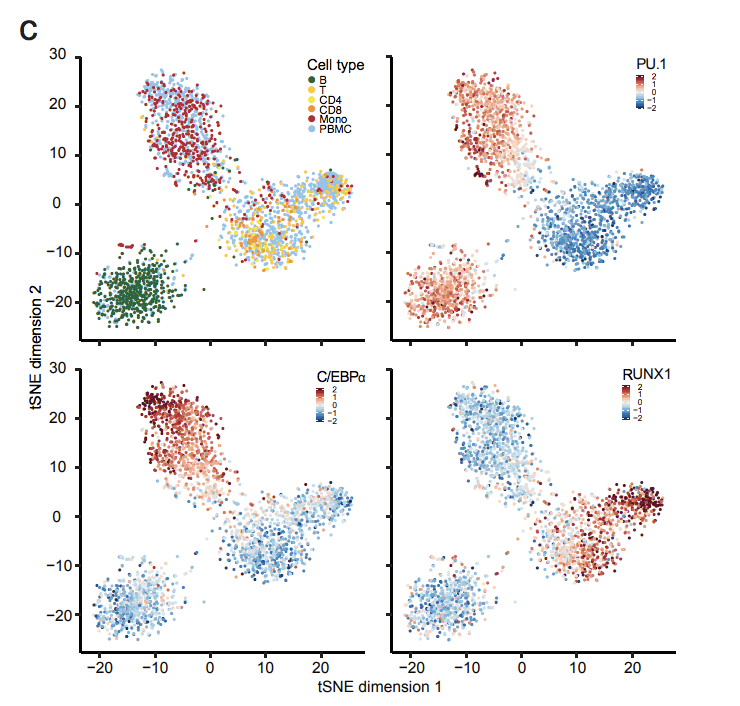
Diversification of DNA binding specificities enabled SREBP transcription regulators to expand the repertoire of cellular functions that they govern in fungi.
Toledo, V. D. O., Puccinelli, R., Fordyce, P.M., and Perez, J.C. PLoS Genetics (in press).
BET-seq: Binding energy topographies revealed by microfluidics and high-throughput sequencing.
Aditham, A.K., Shimko, T.C., and Fordyce, P.M. Methods in Cell Biology - Microfluidics on a Molecular Scale (2018).
Satb1 integrates DNA sequence, shape, motif density and torsional stress to differentially bind targets in nucleosome-dense regions.
Ghosh, R.P., Shi, Q., Yang, L., Reddick, M.P., Nikitina, T., Zhurkin, V.B., Fordyce, P., Stasevich, T.J., Chang, H.Y., Greenleaf, W.J., and Liphardt, J.T., preprint on bioRXiv 10/2018; doi:10.1101/450262. (web)
High-throughput chromatin accessibility profiling at single-cell resolution. Mezger, A., Klemm, S., Mann, I., Brower, K., Mir, A., Bostick, M., Farmer, A., Fordyce, P., Linnarsson, S., & Greenleaf, W.; Nat. Comm. (2018)
(pdf) (web) (bioRXiv doi: 10.1101/310284)
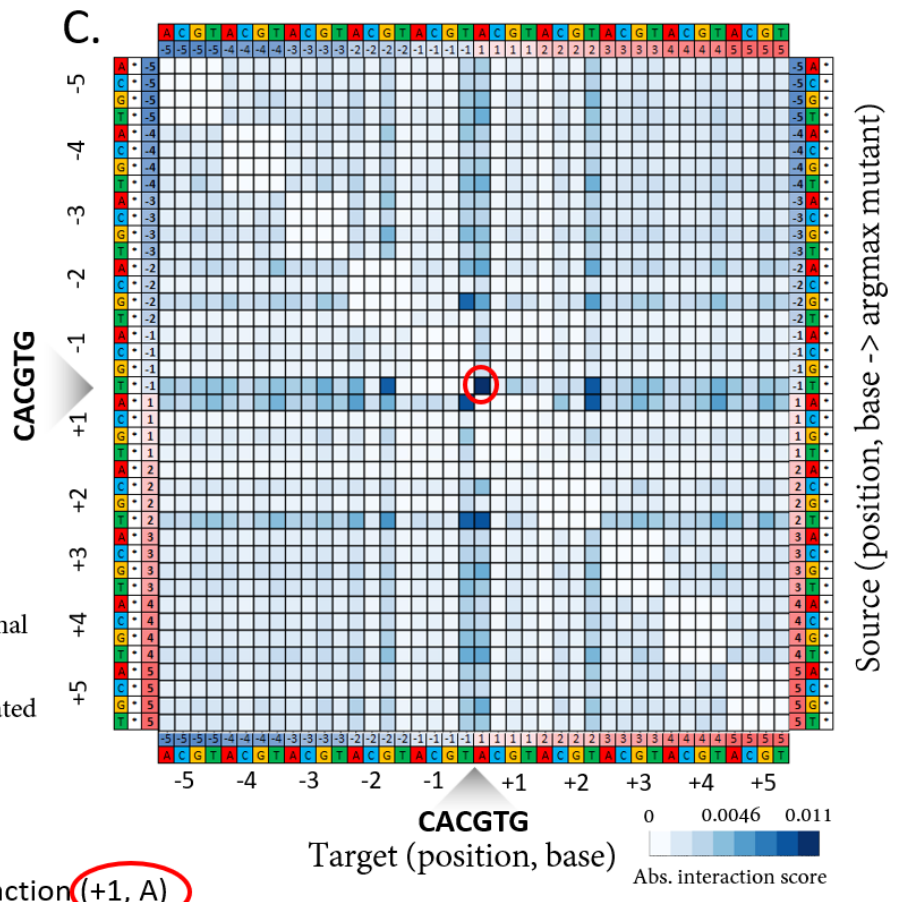
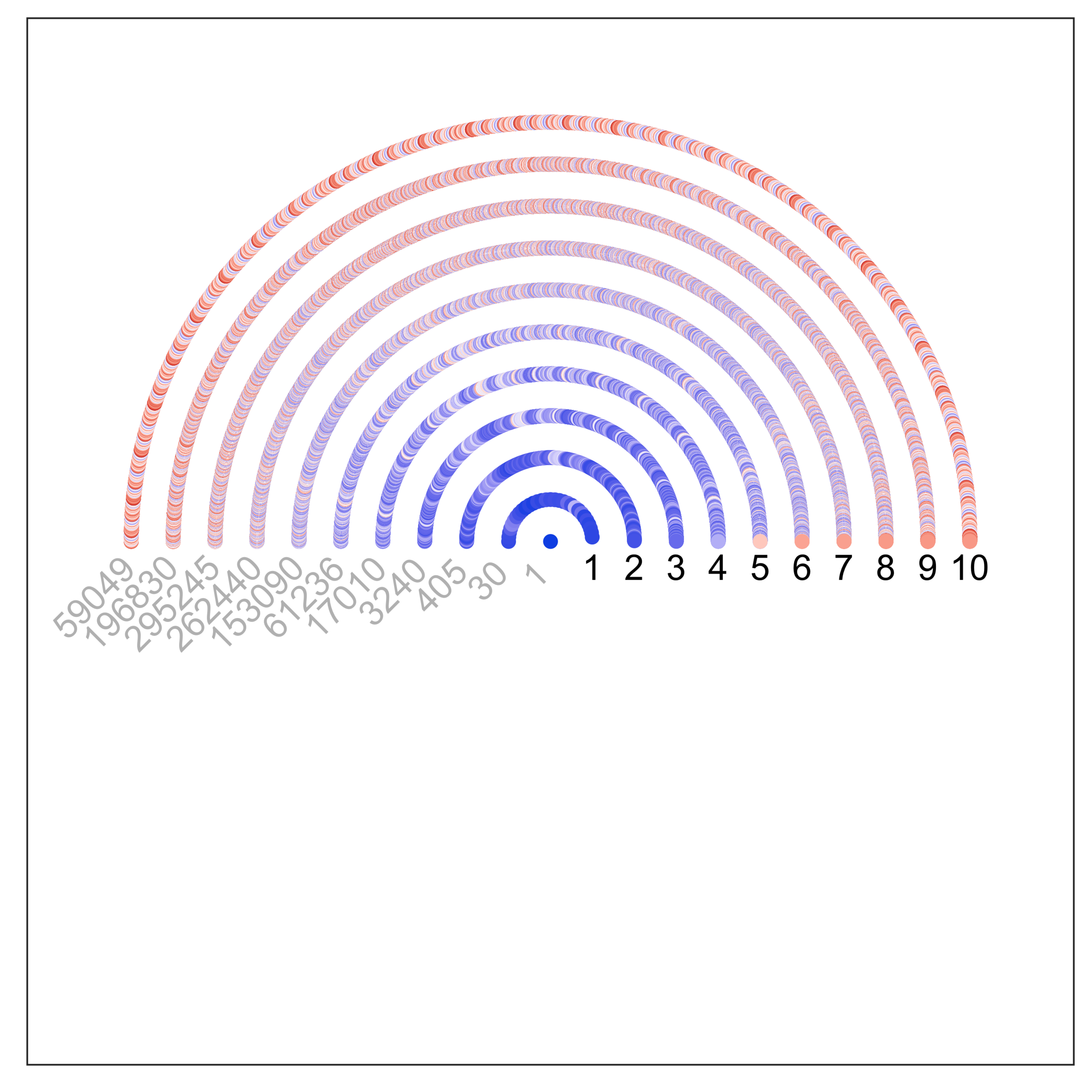
Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Greenside, P.G., Shimko, T., Fordyce, P., & Kundaje, A.; preprint on bioRXiv (04/17/2018); doi: 10.1101/302711 (web)