The Fordyce Lab is focused on developing new instrumentation and assays for making quantitative, systems-scale biophysical measurements of molecular interactions. These tools have many diverse applications, but share a common approach: using techniques and ideas from physics and engineering to improve our understanding of fundamental biological processes.
Current research in the lab is focused on two main areas: using microfluidic tools we have developed to build ground-up quantitative models of how gene expression is regulated, and developing new tools to transform how scientists explore protein-protein interactions.
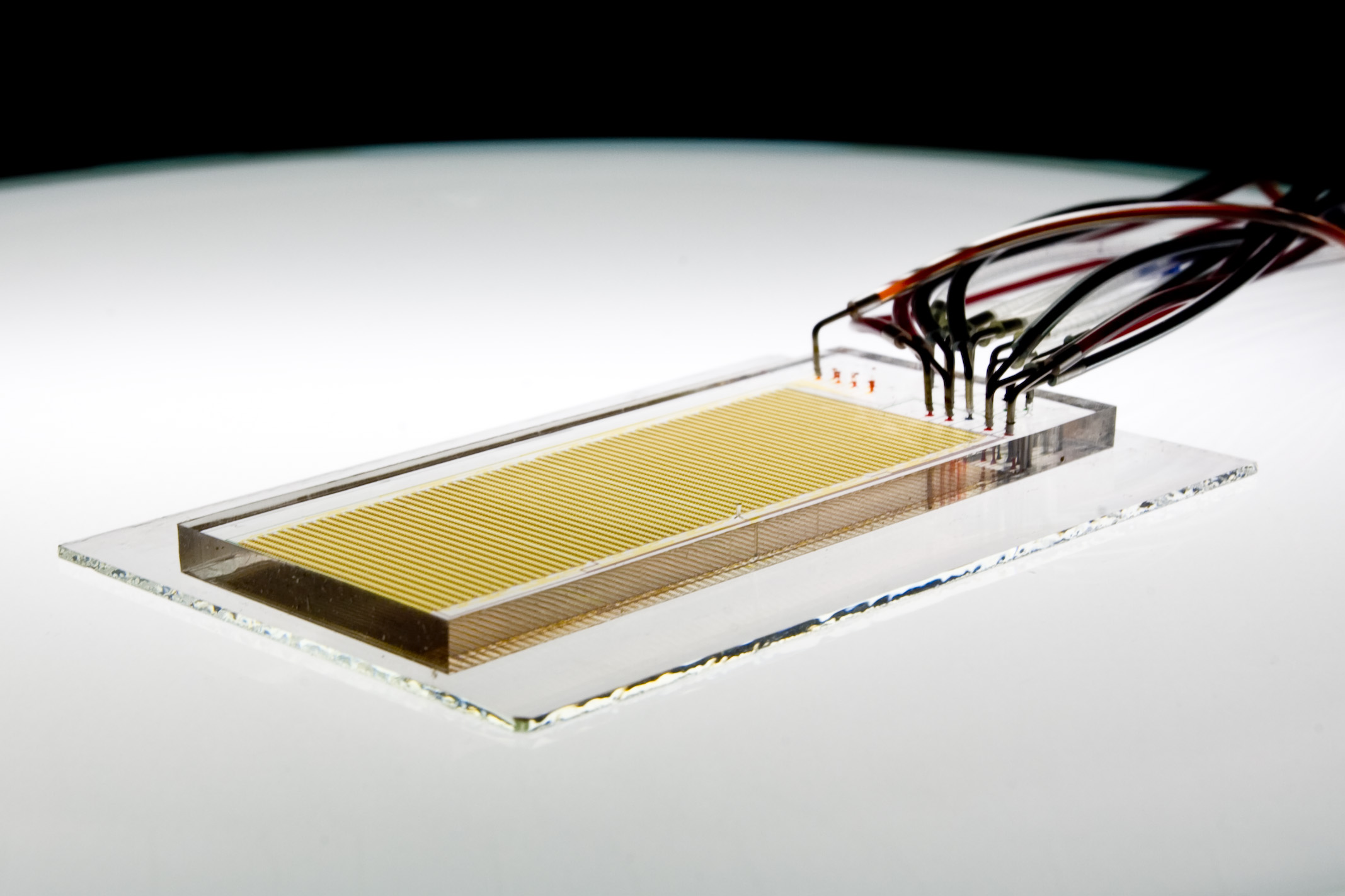
MITOMI 2.0: quantitative measurements of transcription factor binding affinities
Cutting-edge technologies have identified genomic DNA regulatory elements and revealed the binding preferences for many transcription factors. However, our ability to predict in vivo patterns of transcription factor binding from DNA sequence alone remains poor, and measured in vivo binding often cannot explain gene expression patterns, even for well-studied eukaryotic promoters. Improving our understanding of these processes could have far-reaching impacts for revealing how mutations in regulatory elements cause disease and for designing transcriptional circuits for use in synthetic biology. Using a microfluidic technique we have recently developed for making high-throughput, quantitative measurements of transcription factor binding interactions (MITOMI 2.0), we propose to use a "ground-up" approach to reverse engineer transcriptional regulation by systematically adding in components and observing how they influence steady-state occupancies and binding kinetics.
Spectrally encoding beads for biological multiplexing
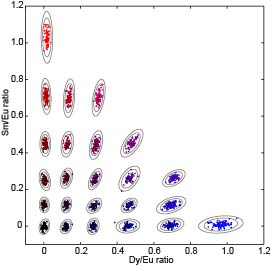
Biological multiplexing allows using very small amounts of samples to test for many different things in parallel. Bead-based multiplexing has many advantages, including flexibility and compatibility with existing equipment and assays, but poses a central challenge: beads must be encoded in some way. We have recently developed new microfluidic methods to produce beads that are spectrally encoded via the ratiometric incorporation of different lanthanide nanonanophosphors. These materials have unique spectral signatures, meaning that beads can later be imaged to "read" the embedded codes. We have previously demonstrated the ability to make up to 82 distinct codes using three lanthanide species (Europium, Samarium, and Dysprosium). We are currently working on expanding our code space to include up to 1,000 distinct spectral codes by using additional lanthanide species and are also working on functionalizing beads for downstream assays.
New microfluidic sorting strategies
We are also currently developing new microfluidic devices that will allow for testing many beads or cells in parallel and then sorting them one-by-one into multiple downstream bins. We hope that these new devices will allow for sorting both beads and cells based on a variety of complex characteristics (such as stiffness or high-resolution imaging).