2021
Revealing enzyme functional architecture via high-throughput microfluidic enzyme kinetics.
Markin, C.J.*, Mokhtari, D.A.*, Sunden, F., Appel, M.J., Akiva, E., Longwell, S.A., Sabatti, C., Herschlag, D.‡, & Fordyce, P.M.‡, Science (2021). (web) (pdf)
• Science Perspective by Baumer & Whitehead.
• Discussion in Nature.
• Stanford News article.
• Chemical & Engineering News article.
Structure-activity mapping of the peptide- and force-dependent landscape of T-cell activation
Feng, Y., Zhao, X., White, A.K., Garcia, K.C., & Fordyce, P.M., bioRxiv (2021).
(web) (pdf)
Fundamentals to function: quantitative and scalable approaches for measuring protein stability.
Atsavapranee, B.*, Stark, C.D.*, Sunden, S., Thompson, S.‡, & Fordyce, P.M.‡. Cell Systems (2021).
(web) (pdf)
2020
MRBLES 2.0: High-throughput generation of chemically functionalized spectrally and magnetically-encoded hydrogel beads using a simple single-layer microfluidic device.
Feng, Y., White, A.K., Hein, J.B., Appel, E.A., & Fordyce, P.M., Microsystems & Nanoengineering (2020).
High-throughput binding affinity measurements for mutations spanning a transcription factor-DNA interface reveal affinity and specificity determinants
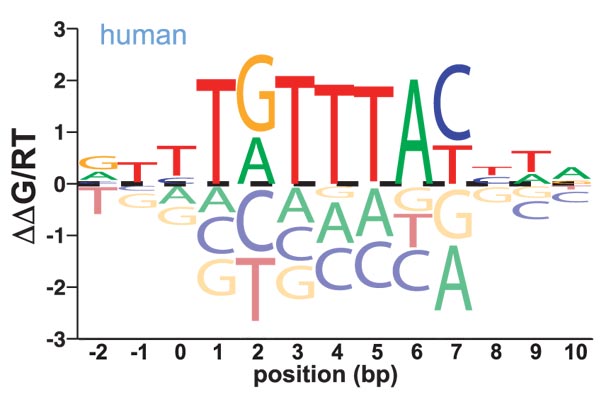
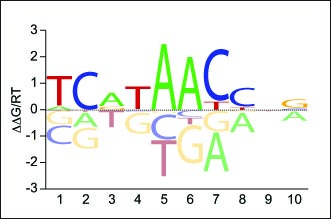
Aditham, A.K., Markin, C.J.*, Mokhtari, D.A.*, DelRosso, N.V., & Fordyce, P.M. Cell Systems (2020).
(pdf) (web)
Double emulsion picoreactors for high-throughput single-cell encapsulation and phenotyping via FACS.
Brower, K.B.*, Khariton, M.*, Suzuki, P.H., Still, C., Kim, G., Calhoun, S.G.K., Qi, L.S., Wang, B.*, & Fordyce, P.M.* Analytical Chemistry (2020).
Optimized double emulsion flow cytometry with high-throughput single-droplet isolation.
Brower, K.B., Carswell-Crumpton, C., Klemm, S., Cruz, B., Kim, G., Calhoun, S.G.K., Nichols, L, & Fordyce, P.M. Lab on a Chip (2020).
2019
Quantitative mapping of protein-peptide affinity landscapes using spectrally encoded beads.
Nguyen, H. Q., Roy, J., Harink, B., Damle, N. P., Latorraca, N. R., Baxter, B. C., Brower, K., Longwell, S., Kortemme, T., Thorn, K. S., Cyert, M. S., Fordyce, P.M eLife 8 (2019): e40499
An open-source software analysis package for Microspheres with Ratiometric Barcode Lanthanide Encoding (MRBLEs)
Harink, B., Nguyen, H., Thorn, K., Fordyce, P. M. PLOS/ONE
2018
Satb1 integrates DNA sequence, shape, motif density and torsional stress to differentially bind targets in nucleosome-dense regions.
Ghosh, R.P., Shi, Q., Yang, L., Reddick, M.P., Nikitina, T., Zhurkin, V.B., Fordyce, P., Stasevich, T.J., Chang, H.Y., Greenleaf, W.J., and Liphardt, J.T., preprint on bioRXiv 10/2018; doi:10.1101/450262. (web)
High-throughput chromatin accessibility profiling at single-cell resolution. Mezger, A., Klemm, S., Mann, I., Brower, K., Mir, A., Bostick, M., Farmer, A., Fordyce, P., Linnarsson, S., & Greenleaf, W.; Nat. Comm. (2018)
(pdf) (web) (bioRXiv doi: 10.1101/310284)
Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Greenside, P.G., Shimko, T., Fordyce, P., & Kundaje, A.; preprint on bioRXiv (04/17/2018); doi: 10.1101/302711 (web)
2017
An open-Source, programmable pneumatic setup for operation and automated control of single-and multi-Layer microfluidic devices. Brower, K., Puccinelli, R., Markin, C., Shimko, T., Longwell, S., Cruz, B., Gomez-Sjoberg, R, Fordyce, P.; HardwareX (2017) (web); preprint on bioRXiv (2017) (web).
OSF repository available here.
Peptide library synthesis on spectrally encoded beads for multiplexed protein/peptide bioassays. Nguyen, H.Q., Brower, K., Harink, B., Baxter, B, Thorn, K., Fordyce, P. Proc. SPIE 10061, Microfluidics, BioMEMS, and Medical Microsystems XV, 100610Z (2017) (web).
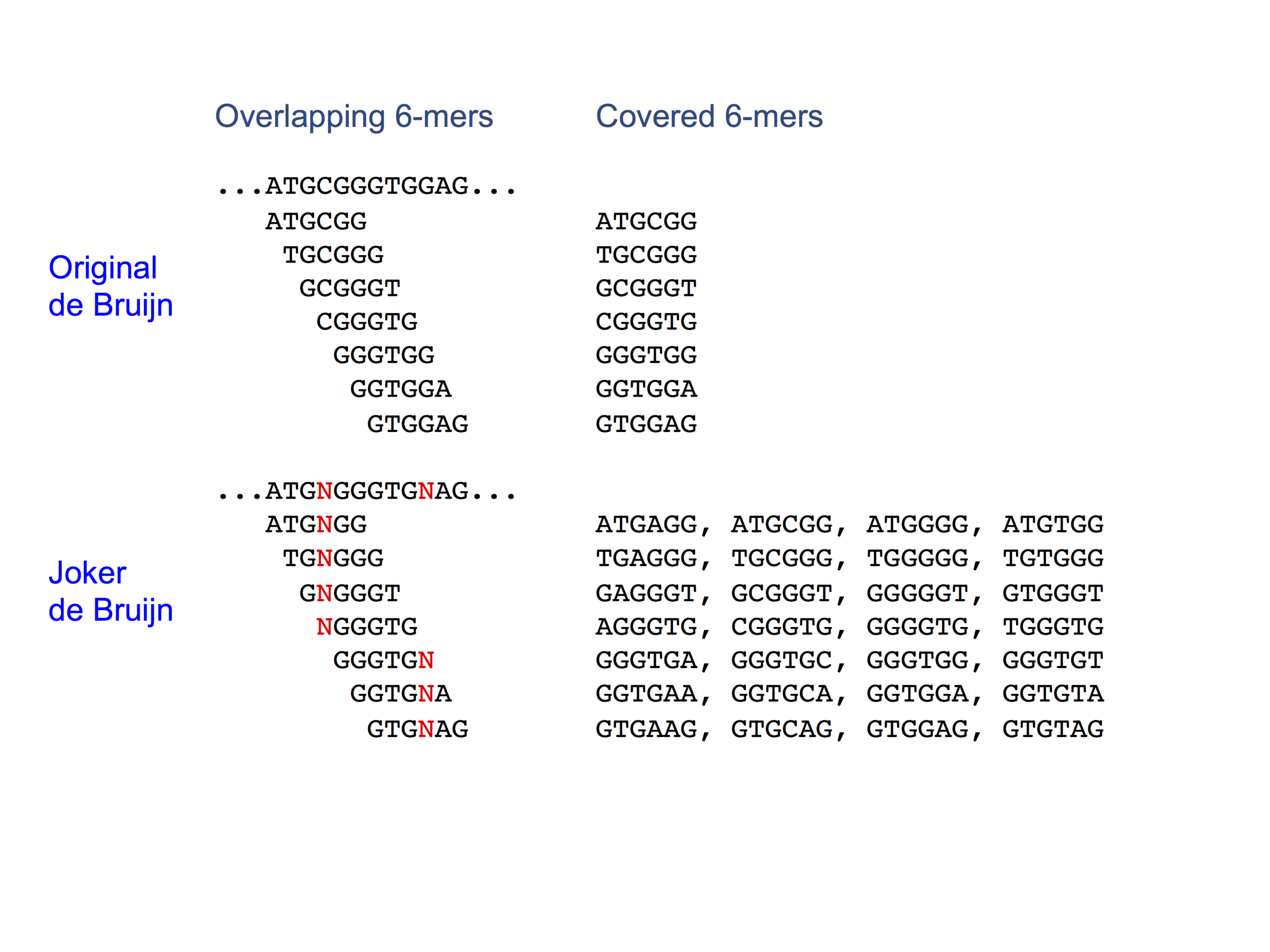
Joker de Bruijn: Sequence libraries to cover all k-mers using joker characters. Ornenstein, Y., Puccinelli, R., Kim, R., Fordyce, P., and Berger, B.; Cell Systems (2017) (web).
2016
2014
2013
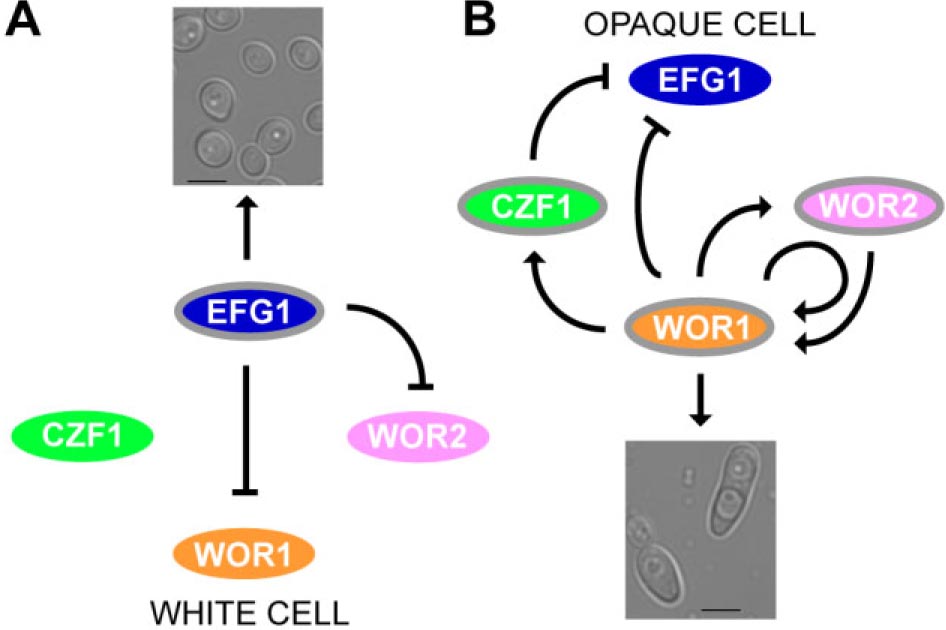
Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains. Lohse, M.B., Hernday, A.D., Fordyce, P.M., Noiman, L., Sorrells, T.R., Hanson-Smith, V., Nobile, C., DeRisi, J.L., & Johnson, A.D. PNAS (2013).
(pdf) (web)
2012
Programmable microfluidic synthesis of spectrally encoded microspheres. Gerver, R.E.*, Gomez-Sjoberg, R.*, Baxter, B.C.*, Thorn, K.S.*, Fordyce, P.M.*, Diaz-Botia, C.A., Helms, B.A., & DeRisi, J.L. Lab on a Chip (2012). (* denotes equal authorship, author order was determined by random draw).










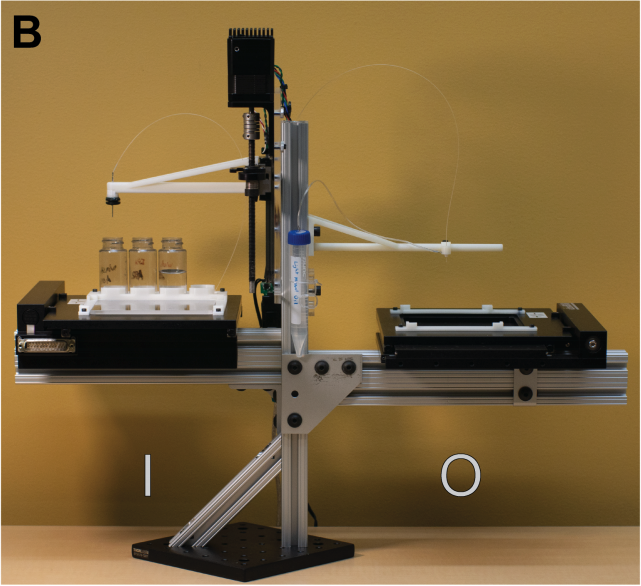
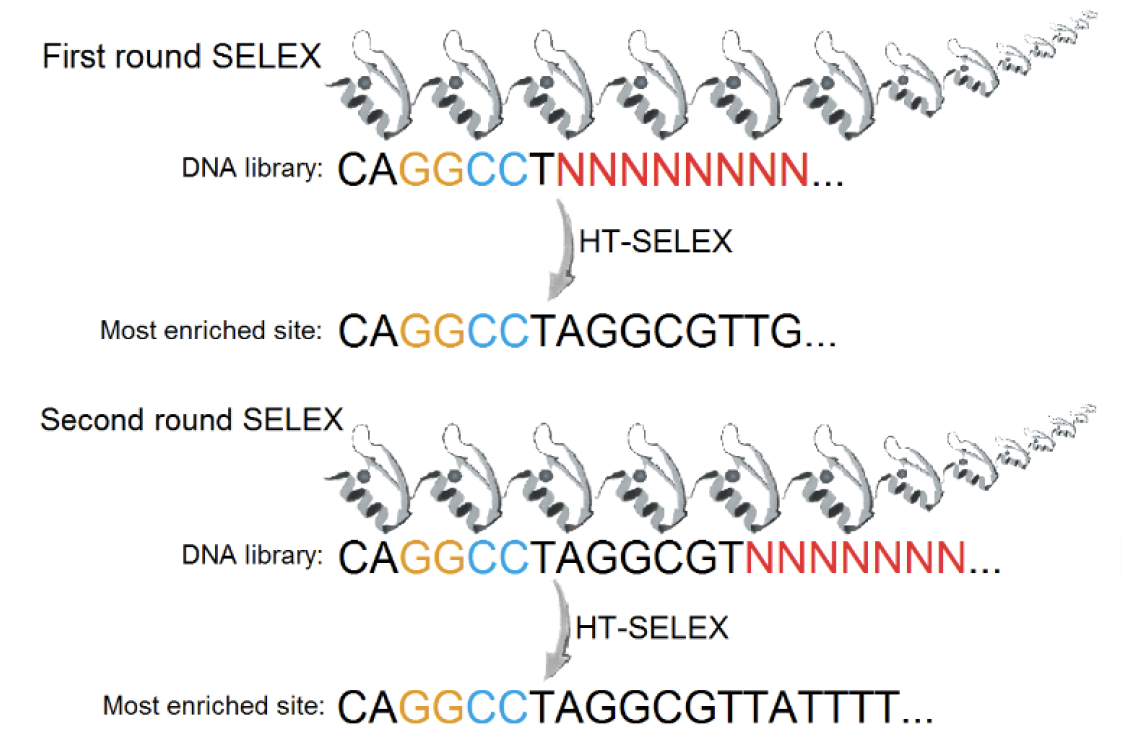
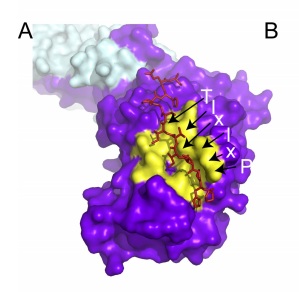
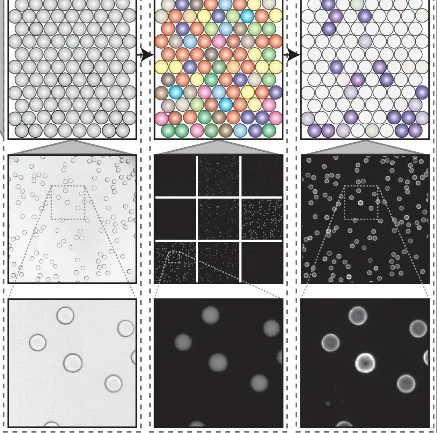
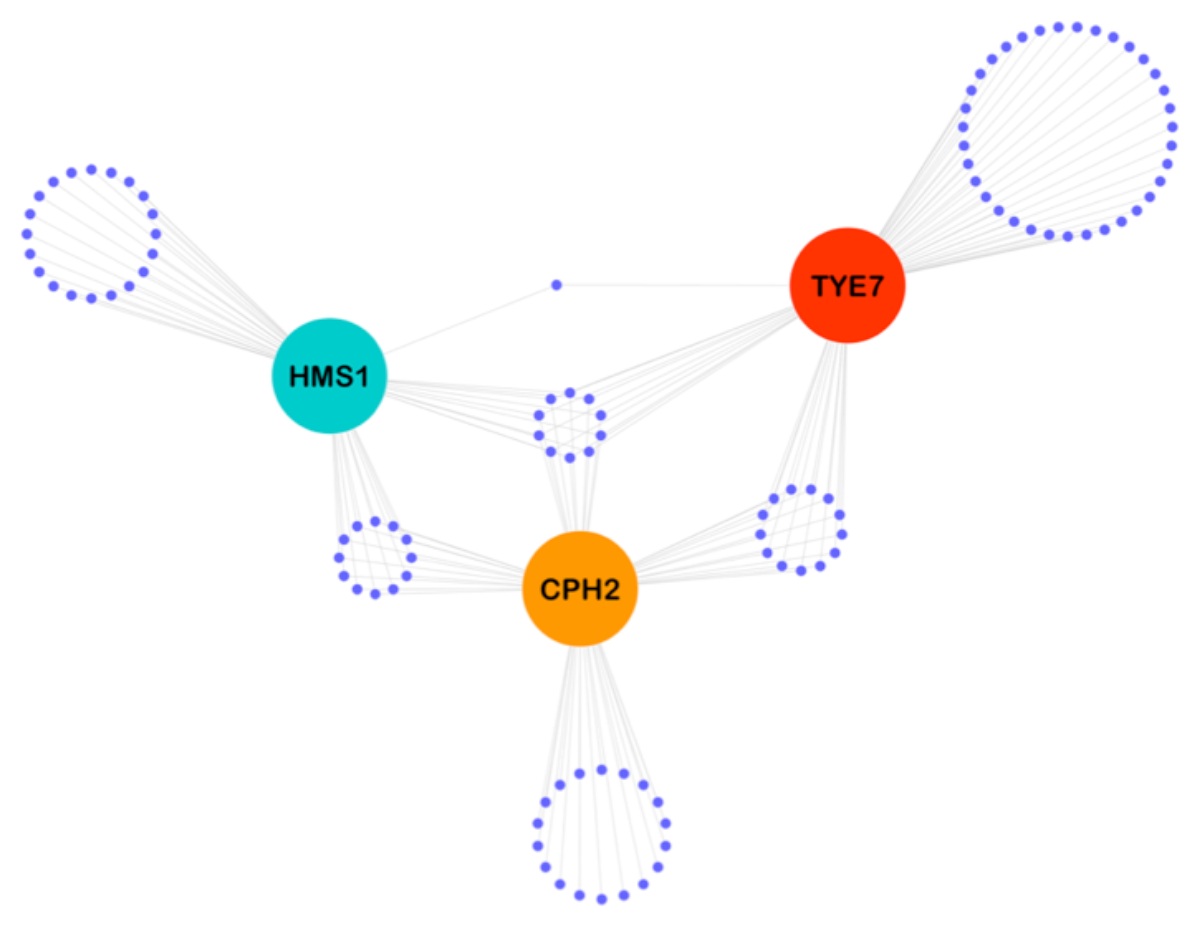
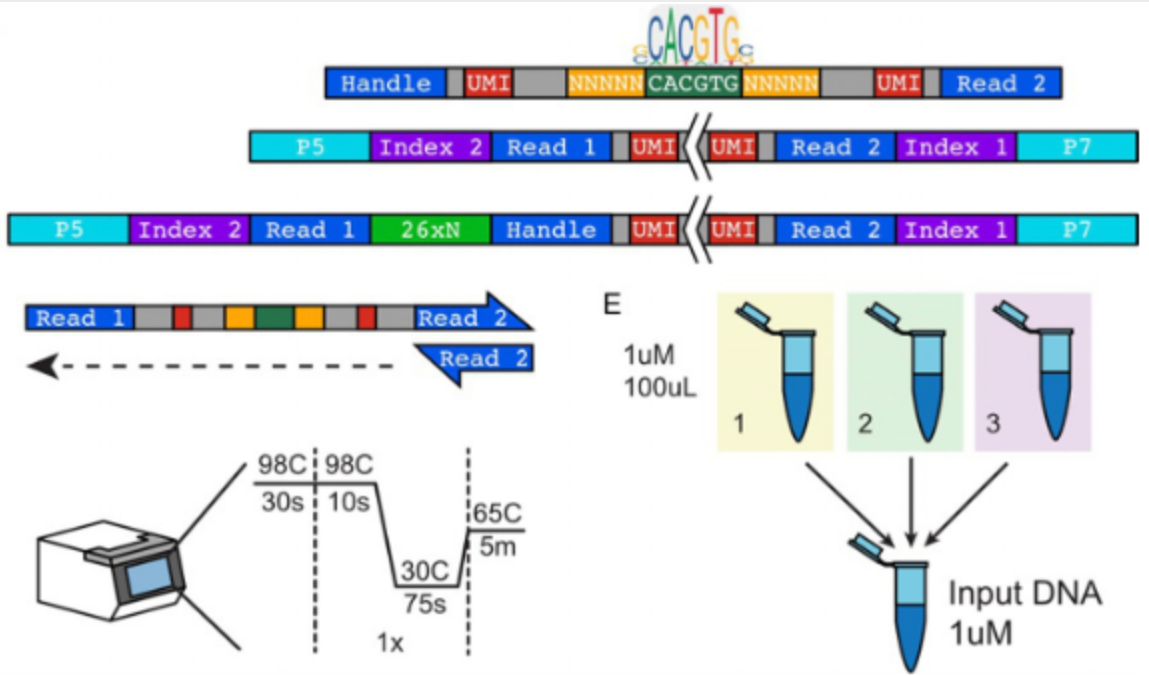
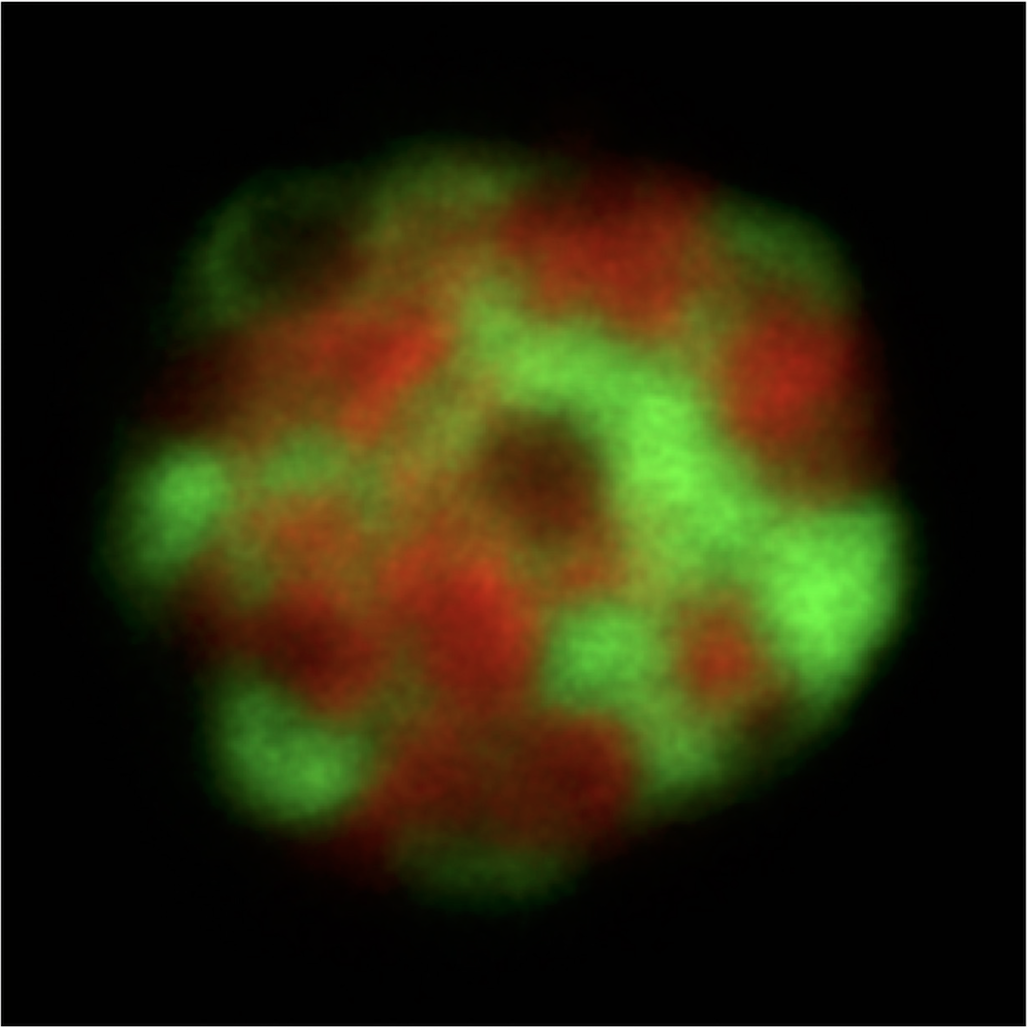
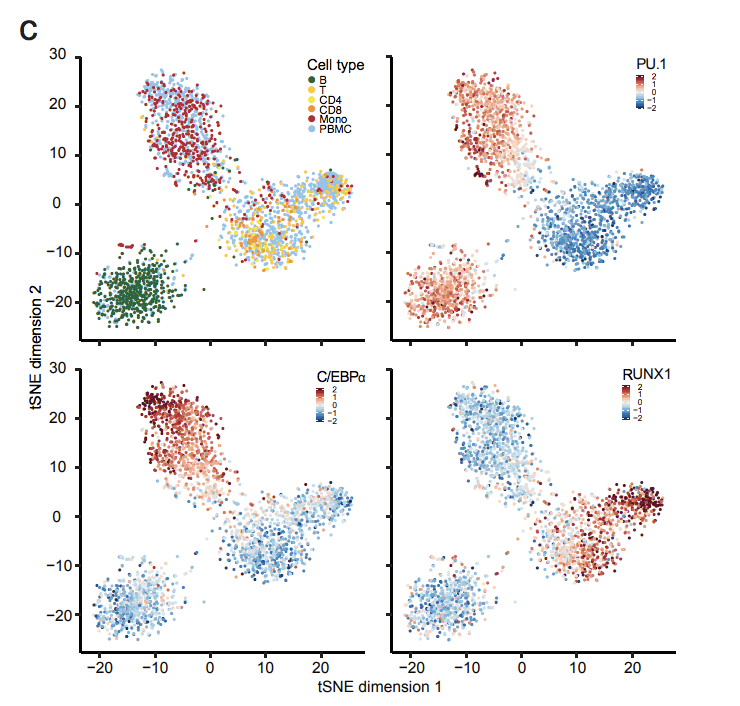
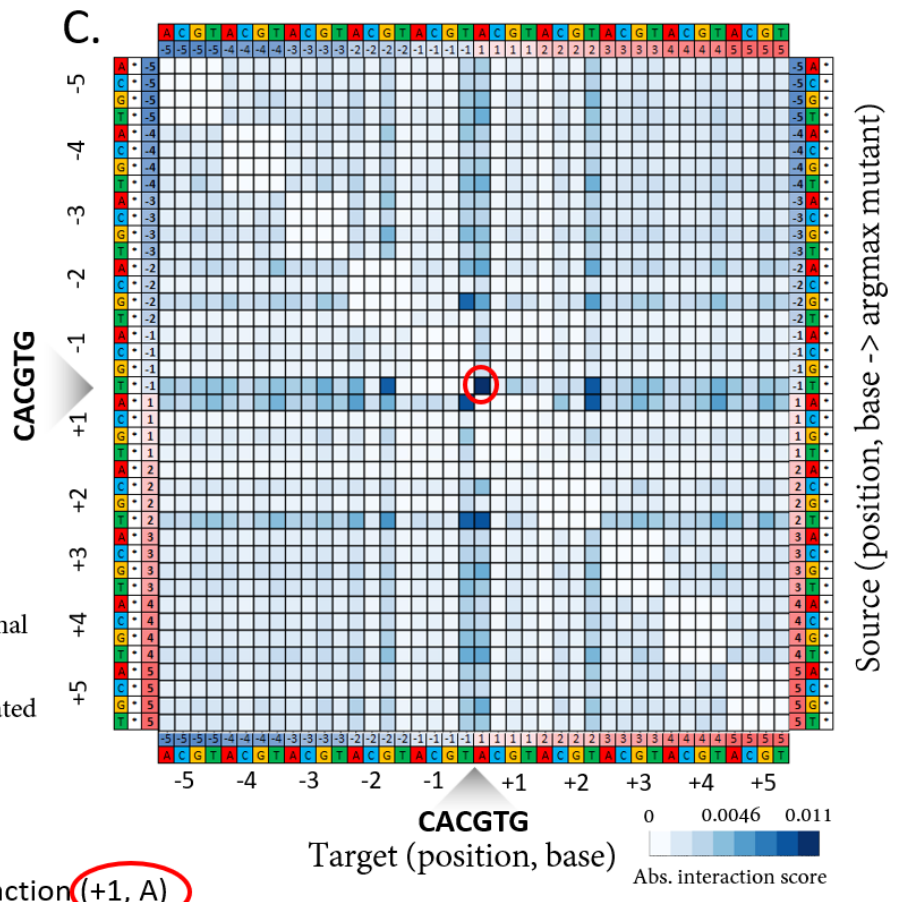
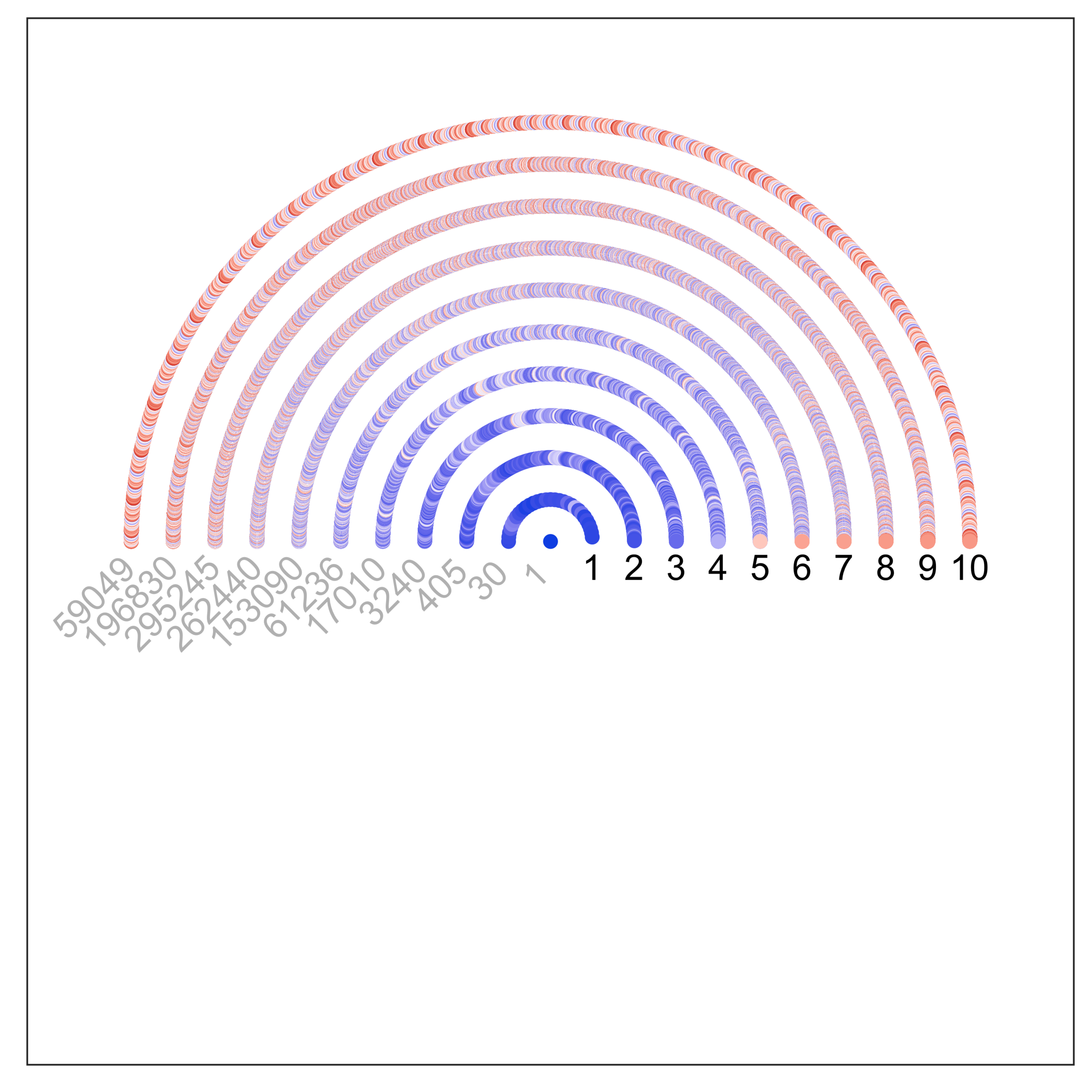
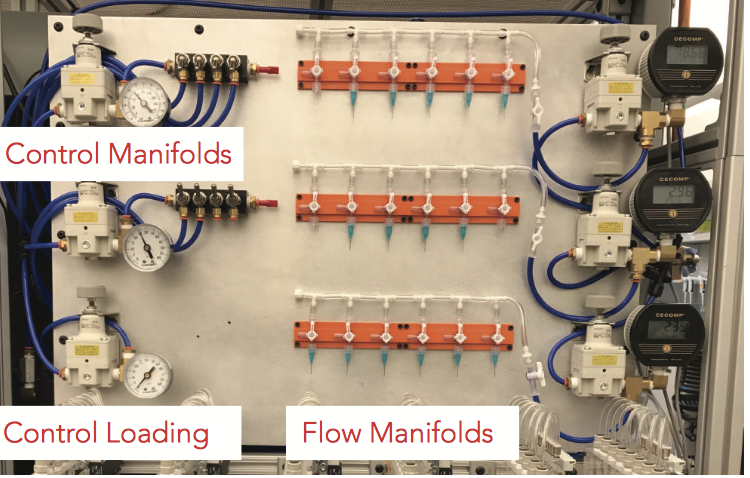




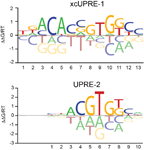
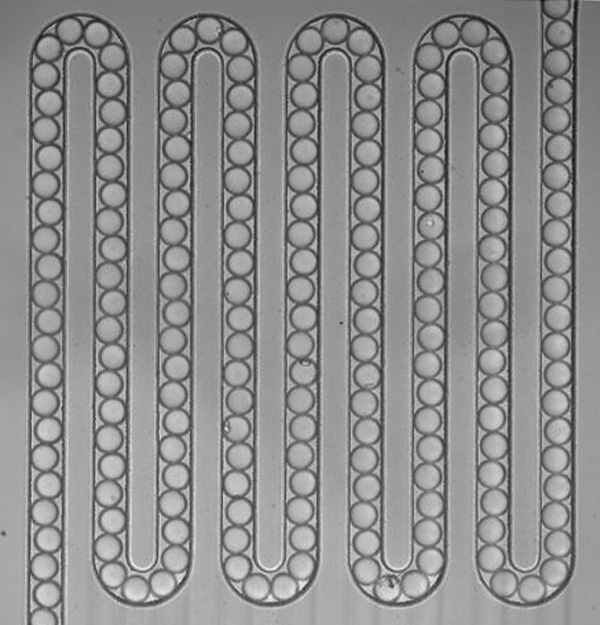
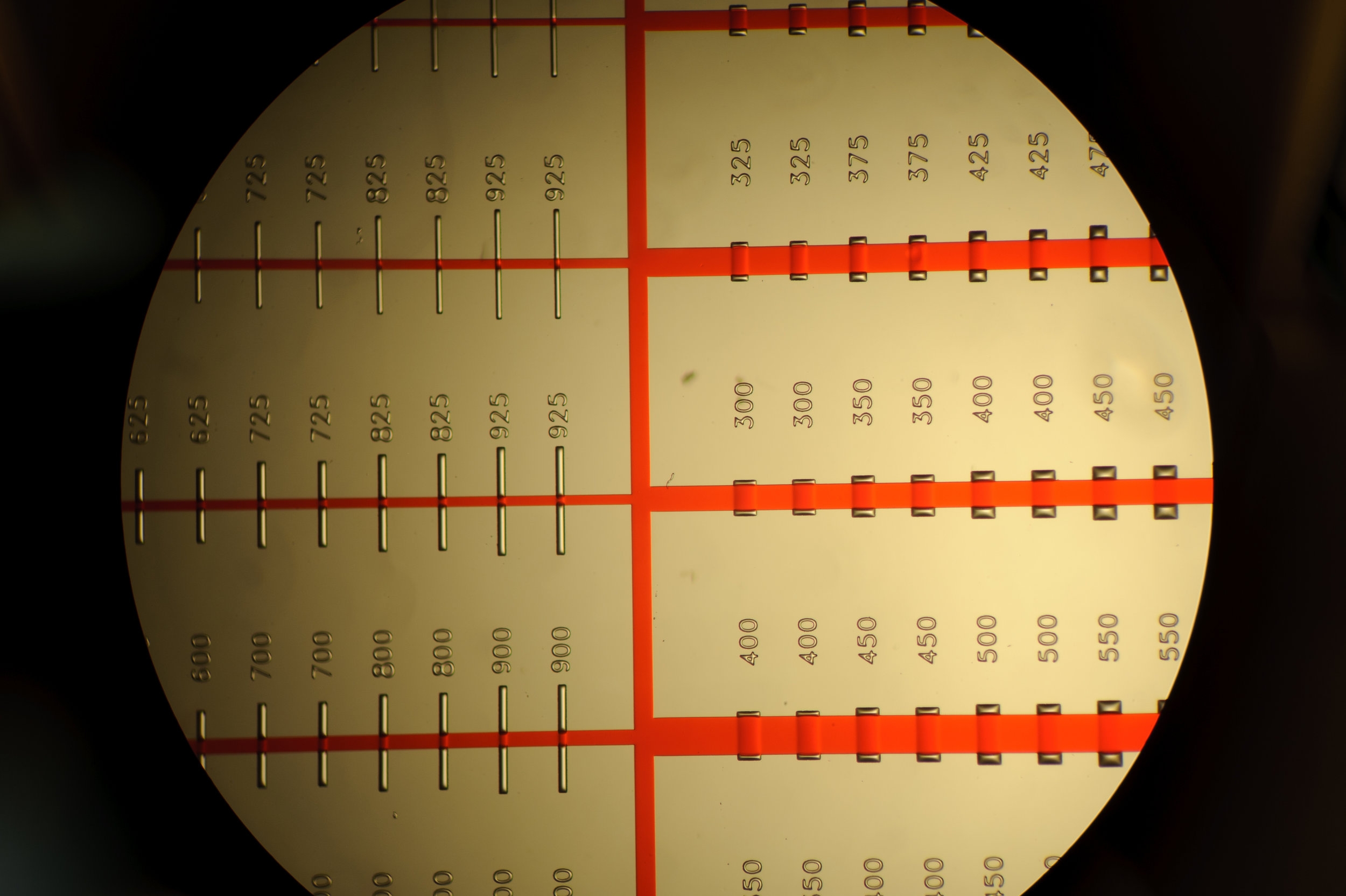

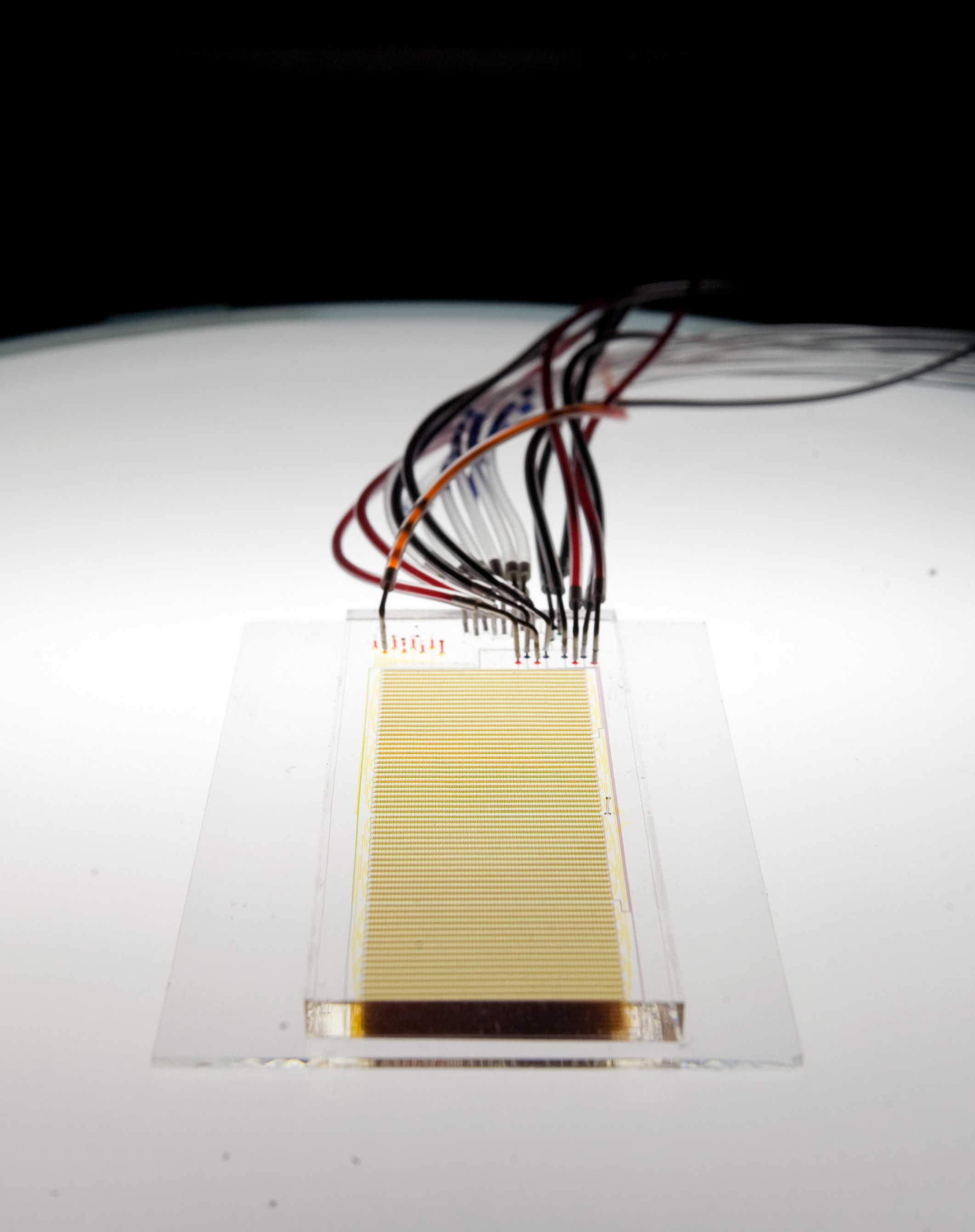




uPIC-M: efficient and scalable preparation of clonal single mutant libraries for high-throughput protein biochemistry.
Appel, M.J., Longwell, S.A., Morri, M., Neff, N., Herschlag, D., & Fordyce, P.M. bioRXiv (2021).
(web) (pdf)